Data Disclaimer & Useful Links
The data used for this database is a high confidence classified subset of larger transposable element sequence data from the Vectorbase Database.
Transposable element data shown was obtained from the Lau Laboratory of Boston University.
Transposable Element Filtering Ma et al. 2020
Methodology to Reduce Redundancy in Mosquito Transposon Family Annotation Lists.
(A) Top: Diagram of the pipeline to condense the AnGam repeats families list, with critical parameters for the BLAT algorithm in the red box. Bottom: Benchmarking results from reducing the AnGam repeats list to 555 repeats families.
(B) Top: Diagram of the pipeline to condense AeAeg repeats families list, with the critical parameters for the MeShClust algorithm in the red cox. Bottom: Benchmarking results for reducing the AeAeg repeats list to 1,242 repeats families.
(C) and (D) The ideogram represents the similar pipeline as the AeAeg pipeline and benchmarking results from reducing repeats families in CuQuin and AeAlbo genomes.

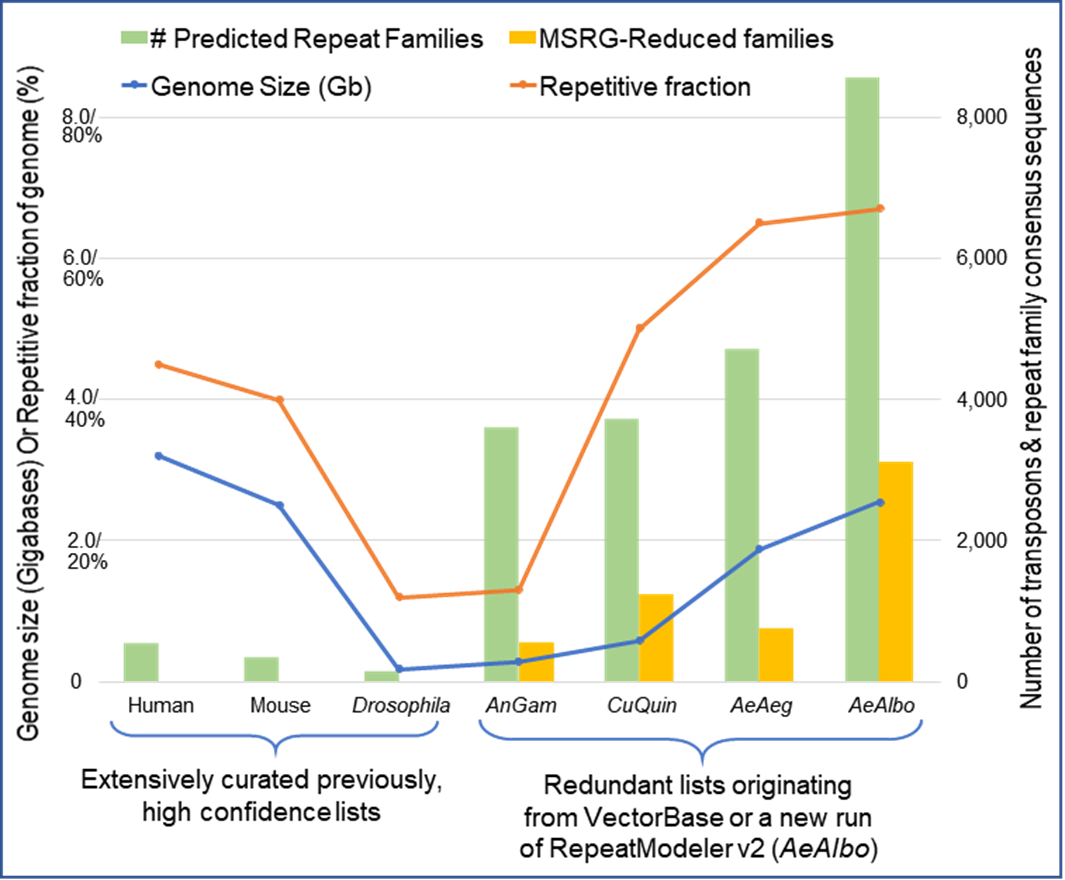
Repetitive Fractions of Genomes Based on Genome Size
Repetitive portions of genomes differ between species. Redudent datasets from VectorBase are a result of RepeatModeler v2 output. Curated genomes are less repetitive.
Recent Publications From Our Collaborators at the Lau Laboratory
