

 |
 |
 |
 |
|||||
Current Research Topics
 |
Our lab's goal is to learn how Piwi proteins and piRNAs evolve during animal evolution to target transposons, viruses and other RNA targets for regulation in germ cell development, animal aging, and responses to virus infection. We utilize Genomics, Bioinformatics, Biochemistry, and Genetic approaches with a wide array of model organisms from Drosophila flies to mosquitos to Xenopus frogs to mammalian brains. |
|
 |
Comparative Genomics of piRNA Clusters in More Strange Vertebrates. We built a comparative genomics pipeline for piRNA-generating genes and intergenic piRNA Clusters in Drosophilids and Mammals. This study revealed the remarkably rapid evolution of piRNA Clusters in animals such that most piRNA loci are species-specific. However, we also uncovered a small group of Eutherian Conserved piRNA Clusters (ECpiC - sounds like "easy-peasy") that may have important functions in mammalian germ cell development. Now we are exploring the comparative genomics of piRNA Clusters in many other cool vertebrate species, from bats to finches to salamanders to pufferfish to sea lamprey. We have all these datasets in hand and are looking for intrepid bioinformaticists to tackle this challenge with us. |
|
|
||
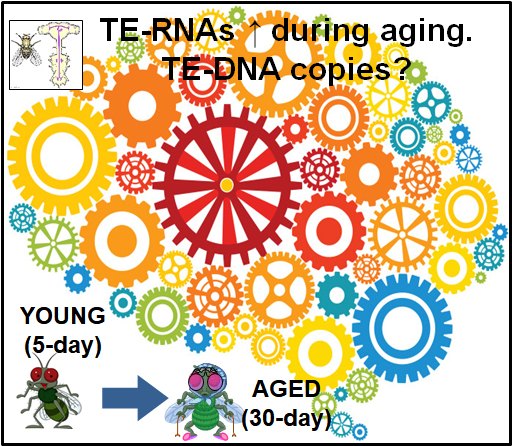 |
Characterizing transposon landscapes during animal aging. Because transposons make up such a large fraction of animal genomes (>50% of the human genome), we carry an immense load of "genetic baggage" that our cells have to keep silent with mechanisms like the RNAi and Piwi pathways. But as our cells age, our chromatin organization and pathways to keep tranposons silent start to break down. We are using our tools like the TIDAL-Fly program to measure transposon landscapes during Drosophila aging. We are now also exploring extending these approaches to mammalian transposons and looking for transposon mRNAs and small RNA signatures to see how they change during aging and neurodegenerative disorders. |
|
|
||
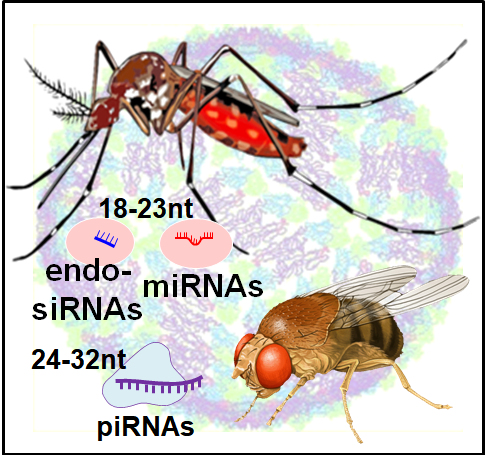 |
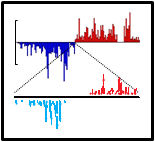
A Mosquito Small RNA Genomics Resource and viral small RNAs. Mosquitoes are relatives of Drosophila fruit flies being part of the Dipteran clade of insects, yet mosquitoes have expanded their Piwi pathway genes and have different compositions of piRNA clusters. Whereas most Drosophila piRNA clusters appear totarget transposons, our analysis of mosquito piRNA clusters indicate few transposons as the targets. These results are reported in a Mosquito Small RNA Genomics resource that will aid our study of how human pathogenic viruses like Dengue and Zika viruses can generate piRNAs. Now we are looking to discover more mosquito piRNA populations from mosquitoes from all over the world to explore the diversity and capacity for persistent viruses in mosquitoes to generate novel piRNAs. |
|
 |
||
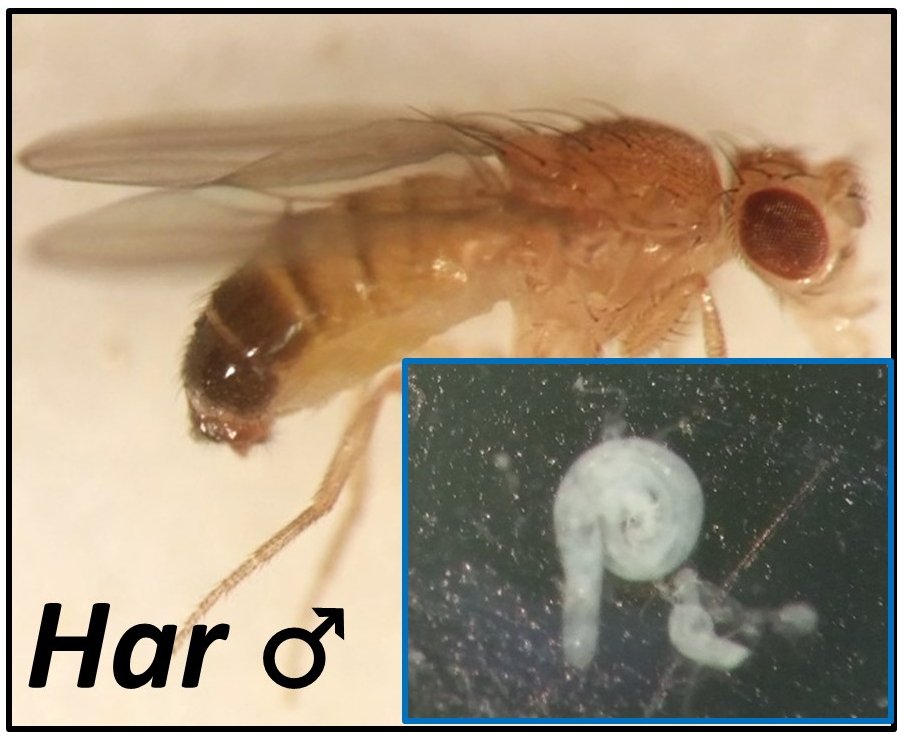 |
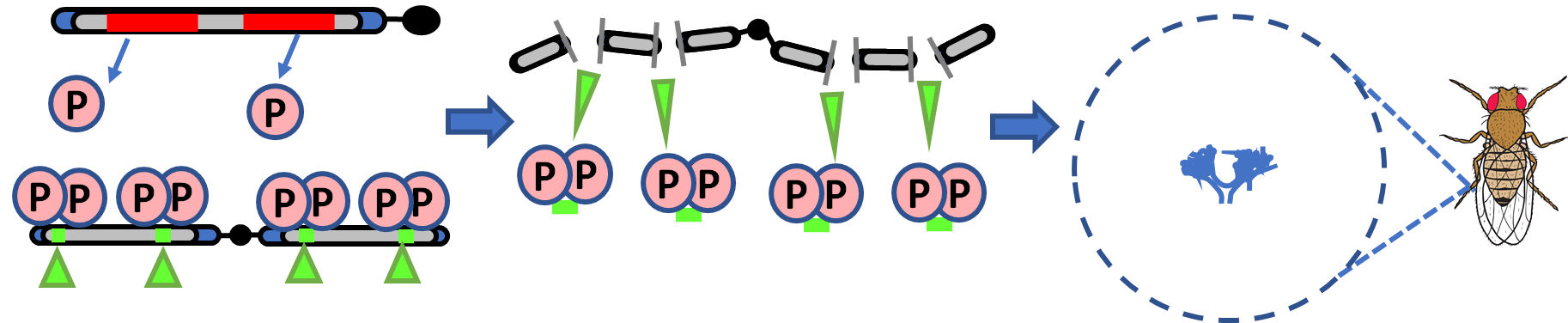
Har-P elements, a hyperactive variant of the P-element transposon in Drosophila. The Harwich strain of Drosophila fruit flies (left) famously has a hyperactive P-element transposon variant that is so actively mobilized that that it causes complete, 100% sterility in daughters when Har males are mated to females lacking piRNAs that would silence the P-element. Now, we want to understand why does this Har-P element damage female germlines more severely than males, and how do piRNA silencing marks on Har-P get passed on from mothers to sons to daughters in a true Drosophila example of transgenerational piRNA-mediated silencing. |
|
 |
||
Interested in studying Small Regulatory RNAs?
See Join the Lab link for more Info.
page updated 20230706
