
 |
 |
 |
 |
|||||
Publications and Protocols
Publications since the lab moved to Boston University Feitosa-Suntheimer F, Zhu Z, Mameli E, Dayama G, Gold AS, Broos-Caldwell A, Troupin A, Rippee-Brooks M, Corley RB, Lau NC, Colpitts TM, Londoño-Renteria B. Dengue Virus-2 Infection Affects Fecundity and Elicits Specific Transcriptional Changes in the Ovaries of Aedes aegypti Mosquitoes. Front Microbiol. 2022 Jun 23;13:886787. doi: 10.3389/fmicb.2022.886787. eCollection 2022. PMID: 35814655. PMCID: PMC9260120 [Journal Link] Henderson C, Brustolin M, Hegde S, Dayama G, Lau N, Hughes GL, Bergey C, Rasgon JL. Transcriptomic and small RNA response to Mayaro virus infection in Anopheles stephensi mosquitoes. PLoS Negl Trop Dis. 2022 Jun 28;16(6):e0010507. doi: 10.1371/journal.pntd.0010507. eCollection 2022 Jun. PMID: 35763539. PMCID: PMC9273063 [Journal Link] Dayama G, Bulekova K, Lau NC. Extending and Running the Mosquito Small RNA Genomics Resource Pipeline. Methods Mol Biol. (2022) ;2509:341-352. doi: 10.1007/978-1-0716-2380-0_20. PMID: 35796973 [Journal Link] Yang N, Srivastav SP, Rahman R, Ma Q, Dayama G, Li S, Chinen M, Lei EP, Rosbash M, Lau NC (2022). Transposable element landscapes in aging Drosophila. PLoS Genet. 2022 Mar 3;18(3):e1010024. doi: 10.1371/journal.pgen.1010024. eCollection 2022 Mar. [Journal Link] Bandyopadhyay S, Douglass J, Kapell S, Khan N, Feitosa-Suntheimer F, Klein JA, Temple J, Brown-Culbertson J, Tavares AH, Saeed M, Lau NC (2021). DNA templates with blocked long 3' end single-stranded overhangs (BL3SSO) promote bona fide Cas9-stimulated homology-directed repair of long transgenes into endogenous gene loci.G3:Genes|Genomes|Genetics (Bethesda). 2021 May 14:jkab169. doi: 10.1093/g3journal/jkab169. PMID: 33989385 [Journal Link] Ma Q, Srivastav SP, Gamez S, Dayama G, Feitosa-Suntheimer F, Patterson EI, Johnson RM, Matson EM, Gold AS, Brackney DE, Connor JH, Colpitts TM, Hughes GL, Rasgon JL, Nolan T, Akbari OS, Lau NC (2021). Diverse Defenses: A Perspective Comparing Dipteran Piwi-piRNA Pathways. Genome Res. 2021 Mar;31(3):512-528. doi: 10.1101/gr.265157.120. Epub 2021 Jan 8. PMCID: PMC7919454 [Open Access Link] Gamez S, Srivastav S, Akbari OS, Lau NC (2020). Diverse Defenses: A Perspective Comparing Dipteran Piwi-piRNA Pathways. Cells. 2020 Sep 27;9(10):2180. doi: 10.3390/cells9102180. PMCID: PMC7601171 [Open Access Link] Zeldich E, Chen CD, Boden E, Howat B, Nasse JS, Zeldich D, Lambert AG, Yuste A, Cherry JD, Mathias RM, Ma Q, Lau NC, McKee AC, Hatzipetros T, Abraham CR. (2019) Klotho Is Neuroprotective in the Superoxide Dismutase (SOD1G93A) Mouse Model of ALS. J Mol Neurosci. 2019 Oct;69(2):264-285. Epub 2019 Jun 27. PMCID: PMC7008951. [Journal link] [PMC Link] Gushchanskaia ES, Esse R, Ma Q, Lau NC, Grishok A. (2019) Interplay between small RNA pathways shapes chromatin landscapes in C. elegans. Nucleic Acids Res. 2019 Jun 20;47(11):5603-5616. l PMCID: PMC6582410. [Open Access Link] Kozeretska IA, Shulha VI, Serga SV, Rozhok AI, Protsenko OV, Lau NC. A rapid change in P-element-induced hybrid dysgenesis status in Ukrainian populations of Drosophila melanogaster. Biol Lett. (2018) Aug;14(8). pii: 20180184. doi: 10.1098/rsbl.2018.0184. [Journal link] [PMC Link] Clark, J.P. Rahman, R., Yang, N., Yang, LY, Lau, NC. Drosophila PAF1 modulates PIWI silencing capacity. Curr Bio. (2017) Sep 11;27(1):1-9. [Journal link] [PMC Link] |
||
 |
||
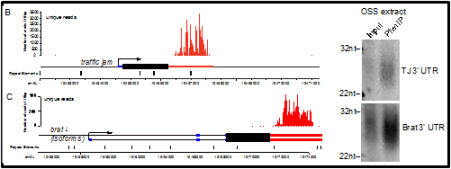
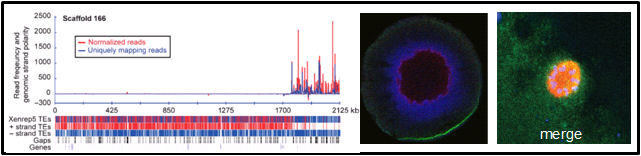
Publications from when the lab was at Brandeis University Toombs, T., Sytnikova, Y., Ang, I., Chirn, GW, Lau, NC*, Blower, MD*. Xenopus Piwi proteins interact with a broad proportion of the oocyte transcriptome. RNA. (2017) Apr;23(4):504-520. doi: 10.1261/rna.058859.116. Epub 2016 Dec 28. *Co-corresponding Author. [Journal link] [PMC Link] Madison-Villar,M., Sun, C., Lau, NC, Settles,M., Mueller, RL. Small RNAs from a big genome: the piRNA pathway and transposable elements in the salamander species Desmognathus fuscus. (2016). J Mol Evol. Oct;83(3-4):126-136. Epub 2016 Oct 14. [Journal link] Rahman, R, Chirn,GW, Kanodia, A, Sytnikova, YA, Brembs, B, Bergman, CM, and Lau, NC. (2015) Unique transposon landscapes are pervasive across Drosophila melanogaster genomes. Nucleic Acids Res. doi: 10.1093/nar/gkv1193 [Open Access Link] Chirn,GW, Rahman, R, Sytnikova, YA, Matts, JA, Zeng, M, Gerlach, D, Yu, M, Berger, B, Kile, BT, and Lau, NC. (2015) Conserved piRNA expression from a distinct set of piRNA cluster loci in Eutherian mammals. PLoS Genetics. doi: 10.1371/journal.pgen.1005652 [Open Access Link] Sytnikova*, YA., Rahman*, R., Chirn, G.W., Post, C., Clark, J., and Lau, N.C. (2014). Transposable element dynamics and PIWI regulation impacts lncRNA and gene expression diversity in Drosophila ovarian cell cultures. Genome Res. Dec;24(12):1977-90. [PDF]. Additional Items For This Paper. Post, C*, Clark*, JP, Sytnikova*, Y., Chirn, G.W., and Lau, N.C. (2014). The capacity of target silencing by the Drosophila Piwi protein and piRNAs. RNA. Dec;20(12):1977-86. [PDF]. Clark, JP, and Lau, N.C. (2014) Piwi proteins and piRNAs step onto the systems biology stage. Adv Exp Med Biol. Systems Biology and Genomics of RNA binding proteins and RNA processing. 2014;825:159-97. doi: 10.1007/978-1-4939-1221-6_5. [PDF] Ow MC, Lau NC, Hall SE. (2014) Small RNA Library Cloning Procedure for Deep Sequencing of Specific Endogenous siRNA Classes in Caenorhabditis elegans. Methods Mol Biol. 1173:59-70. doi: 10.1007/978-1-4939-0931-5_6. [PDF]. Arnold CD, Gerlach D, Spies D, Matts JA, Sytnikova YA, Pagani M, Lau NC, Stark A. (2014). Quantitative genome-wide enhancer activity maps in five Drosophila species reveal the extent of functional enhancer conservation and turnover during cis-regulatory evolution. Nature Genetics, 2014 Jul;46(7):685-92. [PDF]. Ghiretti AE, Moore AR, Brenner RG, Chen LF, West AE, Lau NC, Van Hooser SD, Paradis S. (2014) Rem2 is an activity-dependent negative regulator of dendritic complexity in vivo. J Neurosci. Jan 8;34(2):392-407. PMID: 24403140 PMCID: PMC3870928. [PDF]. Matts, JA*, Sytnikova, Y*, Chirn, G., Igloi, G.L., and Lau, N.C. (2014). Small RNA library construction from minute biological samples. Methods in Molecular Biology. Ed. M. Siomi. 1093:123-36. [PDF]. Zeng, M., Kuzirian M.S., Harper, L., Paradis, S., and Lau, N.C. (2013). Organic small hairpin RNAs (OshR): a Do-It-Yourself platform for transgene-based gene silencing. Methods. 2013 May 23. pii: S1046-2023(13)00148-5. doi: 10.1016/j.ymeth.2013.05.007. [PDF]. Sytnikova, Y, and Lau, N.C. (2013). Does the APC/C mark MIWI and piRNAs for a final farewell? Developmental Cell. Jan 28;24(2):119-20. [PDF]. Hall, SE†, Chirn, G.W., Lau, NC† and Sengupta, P. (2013). RNAi pathways contribute to developmental history-dependent phenotypic plasticity in C. elegans. RNA. Mar;19(3):306-19. [PDF]. Lau NC. (2010) Guiding and guarding gonads: animal germline small RNAs. Int J Biochem Cell Biol. Aug;42(8):1334-47. Epub 2010 Mar 19. [PDF]. Robine N*, Lau NC*†, Balla S*, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. (2009) A broadly conserved pathway generates 3' UTR-directed primary piRNAs. Current Bio. Dec 29;19(24):2066-76. [PDF]. Lau NC, Ohsumi T, Borowsky ML, Kingston RE, Blower MD. (2009) Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO Journal. Oct 7;28(19):2945-58. [PDF]. |
||
 |
||
Earlier Publications Before the Lau lab started Lau NC.Analysis of small endogenous RNAs. Curr Protoc Mol Biol. 2008 Jan;Chapter 26:Unit26.7. [PDF]. Lim LP*, Lau NC*, Weinstein EG*, Abdelhakim AH*, Yekta SO, Rhoades MW, Burge CB, Bartel DP. The MicroRNAs of Caenorhabditis elegans. Genes Dev. 2003 Apr 15;17(8):991-1008. Epub 2003 Apr 02. Landthaler M, Begley U, Lau NC, Shub DA. Two self-splicing group I introns in the ribonucleotide reductase large subunit gene of Staphylococcus aureus phage Twort. Nucleic Acids Res. 2002 May 1;30(9):1935-43. PMCID: PMC113830. Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001 Oct 26;294(5543):858-62 |
||
 |
||
Protocols Small RNA Library Construction for Illumina GAII platform. [PDF]. Quick Q-column Chromatography Isolation of Argonaute and Piwi Complexes. [PDF].
|
page updated 20230706